1.Load packages we will use
download co2 emissions per capita from Our world in Data into the directory for the post.
Assign the location of the file to
file_csv. The data should be in the same directory as this file. -Read the data into R and assignemissions
file_csv <- here("_posts","2021-02-16-reading-and-writting-data","co-emissions-per-capita.csv")
emissions <- read_csv(file_csv)
- show the first 10 rows (observation of)
emissions
emissions
# A tibble: 22,383 x 4
Entity Code Year `Per capita CO2 emissions`
<chr> <chr> <dbl> <dbl>
1 Afghanistan AFG 1949 0.00191
2 Afghanistan AFG 1950 0.0109
3 Afghanistan AFG 1951 0.0117
4 Afghanistan AFG 1952 0.0115
5 Afghanistan AFG 1953 0.0132
6 Afghanistan AFG 1954 0.0130
7 Afghanistan AFG 1955 0.0186
8 Afghanistan AFG 1956 0.0218
9 Afghanistan AFG 1957 0.0343
10 Afghanistan AFG 1958 0.0380
# … with 22,373 more rows5.start with emissions data THEN - Use clean_names from the janitor package to make easier to work with - assign the output totidy_emissions - show the first 10 rows of tidy_emission
tidy_emissions <- emissions %>%
clean_names()
tidy_emissions
# A tibble: 22,383 x 4
entity code year per_capita_co2_emissions
<chr> <chr> <dbl> <dbl>
1 Afghanistan AFG 1949 0.00191
2 Afghanistan AFG 1950 0.0109
3 Afghanistan AFG 1951 0.0117
4 Afghanistan AFG 1952 0.0115
5 Afghanistan AFG 1953 0.0132
6 Afghanistan AFG 1954 0.0130
7 Afghanistan AFG 1955 0.0186
8 Afghanistan AFG 1956 0.0218
9 Afghanistan AFG 1957 0.0343
10 Afghanistan AFG 1958 0.0380
# … with 22,373 more rows6 start with the tidy_emissions THEN -use filter to extract rows with years == 2000 -use skim to calculate the descriptive statistics
| Name | Piped data |
| Number of rows | 219 |
| Number of columns | 4 |
| _______________________ | |
| Column type frequency: | |
| character | 2 |
| numeric | 2 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| entity | 0 | 1.00 | 4 | 32 | 0 | 219 | 0 |
| code | 12 | 0.95 | 3 | 8 | 0 | 207 | 0 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| year | 0 | 1 | 2000.00 | 0.00 | 2e+03 | 2000.00 | 2000.00 | 2000.00 | 2000.00 | ▁▁▇▁▁ |
| per_capita_co2_emissions | 0 | 1 | 5.06 | 6.74 | 2e-02 | 0.71 | 2.82 | 7.97 | 58.39 | ▇▁▁▁▁ |
- 13 observations have a missing code. How are these observations different? -start with
tidy_emissionsthen extract rows withyear==2000and are missing a code# A tibble: 12 x 4 entity code year per_capita_co2_emissions <chr> <chr> <dbl> <dbl> 1 Africa <NA> 2000 1.11 2 Asia <NA> 2000 2.40 3 Asia (excl. China & India) <NA> 2000 3.35 4 EU-27 <NA> 2000 8.46 5 EU-28 <NA> 2000 8.61 6 Europe <NA> 2000 8.48 7 Europe (excl. EU-27) <NA> 2000 8.47 8 Europe (excl. EU-28) <NA> 2000 8.19 9 North America <NA> 2000 14.6 10 North America (excl. USA) <NA> 2000 5.39 11 Oceania <NA> 2000 12.6 12 South America <NA> 2000 2.32- Start with the tidy_emissions THEN -use
filterto extract rows with year==2000 and without missing codes THEN -useselectto drop theyearvariable THEN -userenameto change the variableentitytocountry-assign the output toemissions_2000- Which 15 countries have the highest
per_capita_co2_emissions? -start withemissions_2000THEN -useslice_maxto extract then 15 rows with theper_caita_co2_emissionsassign the output tomax_15_emittersmax_15_emitters <- emissions_2000 %>% slice_max(per_capita_co2_emissions,n=15)- which 15 countries have the lowest
per_capita_co2_emissions? -start withemissions_2000THEN -useslice_minto extract the 15 rows with the lowest values -assign the outputmin_15_emittersmin_15_emitters <- emissions_2000 %>% slice_min(per_capita_co2_emissions,n=15)11.Use
bind_rowsto bind together themax_15_emittersandmin_15_emitters- assign the output tomax_min_15max_min_15 <- bind_rows(max_15_emitters,min_15_emitters)- export
max_min_15to 3 file formats13 Read the 3 file formats into Rmax_min_15 %>% write_csv("max_min_15.csv")#comma-separated values. max_min_15 %>% write_tsv("max_min_15.tsv")#tab separated. max_min_15 %>% write_delim("max_min_15.psv", delim="|")#pipe-separated.14 usemax_min_15_csv <-read_csv("max_min_15.csv")#comma-separated values. max_min_15_tsv <- read_tsv("max_min_15.tsv")#tab separated. max_min_15_psv<- read_delim("max_min_15.psv", delim="|")#pipe-separated.setdiffto check for any differences amongmax_min_15_csv,max_min_15_tsvandmax_min_15_psvsetdiff(max_min_15_csv, max_min_15_tsv, max_min_15_psv)# A tibble: 0 x 3 # … with 3 variables: country <chr>, code <chr>, # per_capita_co2_emissions <dbl>Are there any differences?
15 readercountryinmax_min_151for plotting and assign to max_min_15_plot_data -start withemissions_2000THEN -usemutateto reordercountryaccordingper_capital_co2_emissionsmax_min_15_plot_data <- max_min_15 %>% mutate(country=reorder(country,per_capita_co2_emissions))- Plot
max_min_15_plot_dataggplot(data=max_min_15_plot_data, mapping = aes(per_capita_co2_emissions, country)) + geom_col() + labs(title = "the top 15 and bottom 15 per capita CO2 emissions", x=NULL, Y=NULL)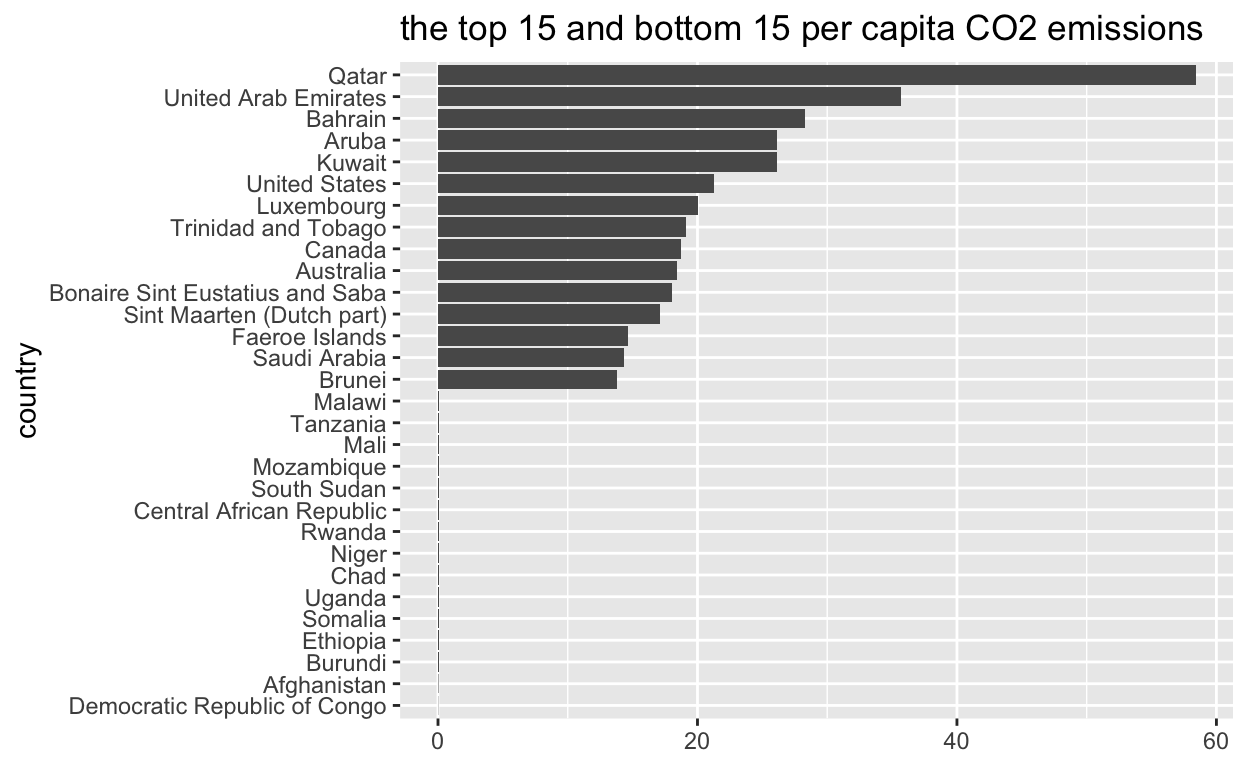
- Save the plot directory with this post
ggsave(filename = "preview.png", path = here("_posts","2021-02-16-reading-and-writting-data"))18 add preview.png to yaml chuck at the top of this file
preview: preview.png
- Save the plot directory with this post
- Plot
- export
- which 15 countries have the lowest
- Which 15 countries have the highest
- Start with the tidy_emissions THEN -use